Our research aims to understand how genetics contributes to the variation between patients in their disease severity and response to treatments. Understanding this variation is vital for improving the success rate of clinical trials and providing a precision medicine approach for treating disease. Our strategy is to not only sequence a patient’s DNA, but also to measure the activity of their genes through RNA-sequencing.
We focus on diseases that involve systemic inflammation such as sepsis and systemic lupus erythematosus (SLE). For both of these diseases, patients present with variable clinical features making diagnosis and treatment challenging. We apply sophisticated analytical methods to analyse functional genomics data generated from cohorts of hundreds of patients and integrate this with clinical information.
Patient stratification from transcriptomic profiling
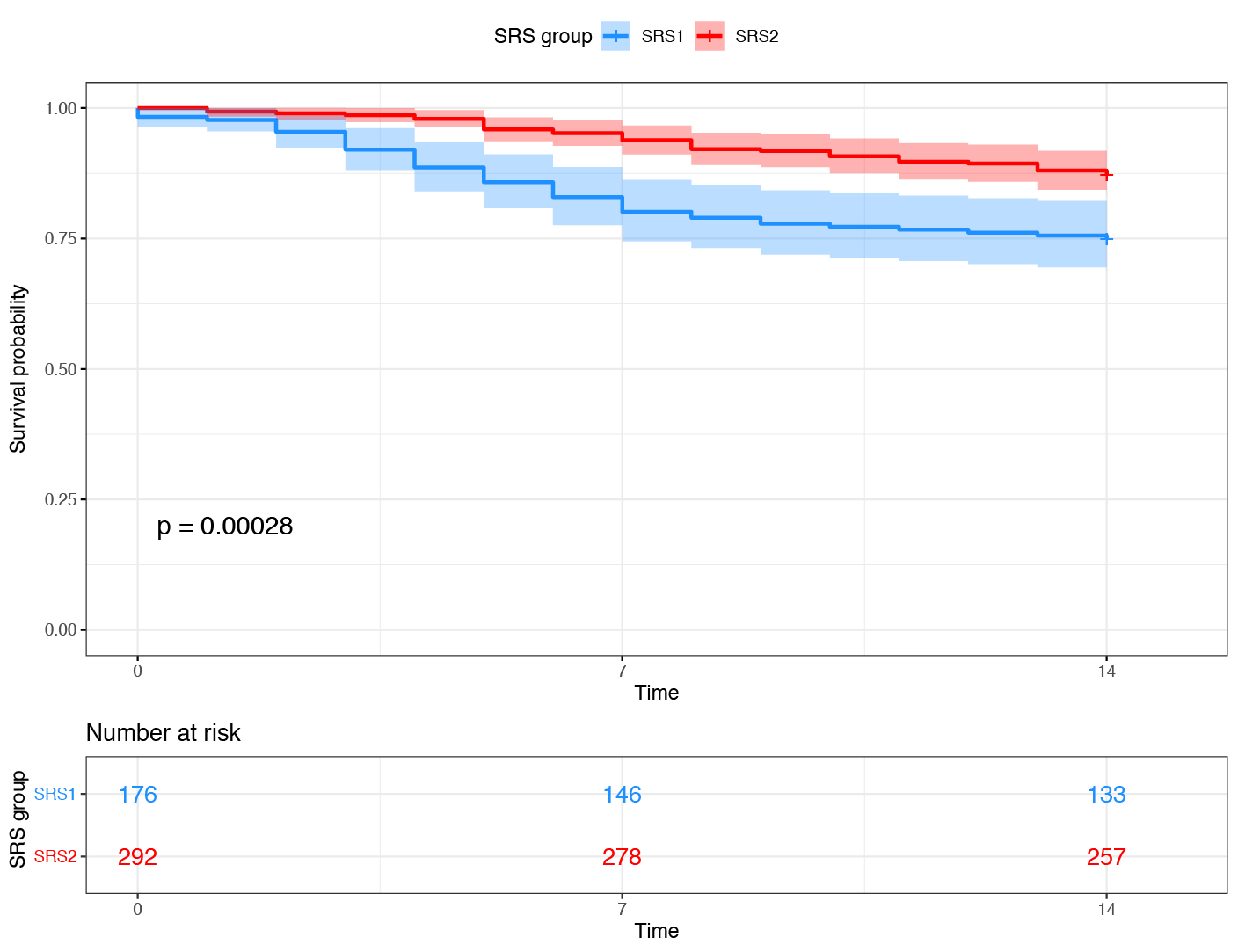
Right: Patients with sepsis due to Community Acquired Pneumonia or Faecal Peritonitis were assigned to Sepsis Response Signature (SRS) endotype, based on the expression of a small number of genes. The SRS1 endotype was associated with stronger features of T cell exhaustion, endotoxin tolerance and immunosuppression, and had a higher mortality rate (blue subgroup in Kaplan-Meier plot showing survival rate over the first 14 days in ICU).
Role of regulatory DNA variants in patient heterogeneity
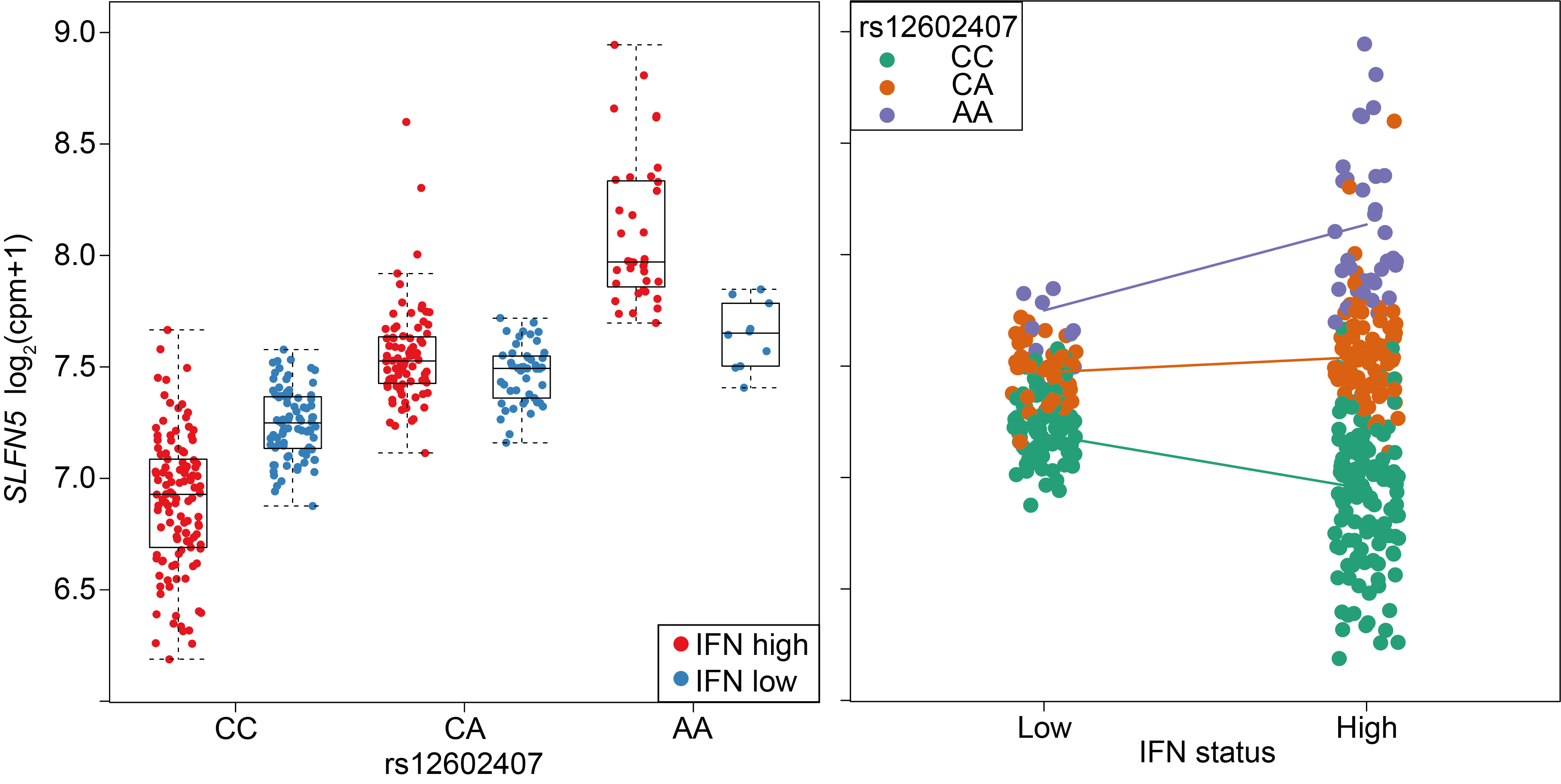 A cohort of patients from a systemic lupus erythematosus clinical trial. An interaction between IFN status and the SLFN5 eQTL is plotted.
Left: with respect to rs12602407 genotype.
Right: with respect to IFN status of the sample.
A cohort of patients from a systemic lupus erythematosus clinical trial. An interaction between IFN status and the SLFN5 eQTL is plotted.
Left: with respect to rs12602407 genotype.
Right: with respect to IFN status of the sample.
Drug target prioritisation from single-cell eQTL mapping
SLE is an autoimmune disease that can affect virtually any organ system in the body. This clinical heterogeneity has made the development of new drugs for treatment very challenging. To enable the development of new therapies, we are interested in better understanding the molecular mechanisms contributing to the disease pathophysiology. Through Open Targets, we are generating single-cell RNA-sequencing for a cohort of SLE patients. This will allow us to map eQTL at the single-cell level and gain mechanistic insights that can inform novel target identification for future drugs.